Hematopoietic stem cell (HSC) transplantation is the only stem cell therapy in routine clinical use, and it is also the cell type that gives rise to most blood cancers. We use single cell biology and genetics to understand how hematopoietic stem cells normally sustain blood formation, and how this process is altered during ageing and when leukemia develops.

Each day the human body generates >500 billion blood cells of more than 10 different types from a few million HSCs. This occurs through a series of progenitor cells, and is sustained by a complex network of stromal cells that both support HSC maintenance and promote the survival and proliferation of differentiating progenitor cells. Our overall aim is to generate an accurate and comprehensive model of hematopoiesis that describes the nature and properties of HSCs, the cellular pathways and molecular mechanisms that govern their differentiation, and the cellular environment that supports this highly efficient process.
The hematopoietic stem cell pool has the capacity to generate erythrocytes, platelets, myeloid and lymphoid cell types in a continuous manner for the entire lifespan of the organism. Until recently it was thought that all blood cell types were produced by all HSCs. By transplanting single murine HSCs we have shown that HSCs exist that produce only a subset of hematopoietic cell types, and in particular that HSCs exist which are biased towards platelet production (Sanjuan-Pla et al., 2013). These HSCs are able to respond to a lack of platelets by expanding to replace the lost platelets. We are currently working to expand this concept to human HSCs, and to understand the molecular mechanisms by which HSCs with selective lineage output are specified, and how they contribute to the increases in blood cell production in emergency situations such as anemia and infection (Grover et al., 2014), and how they change during ageing (Grover et al., 2016).
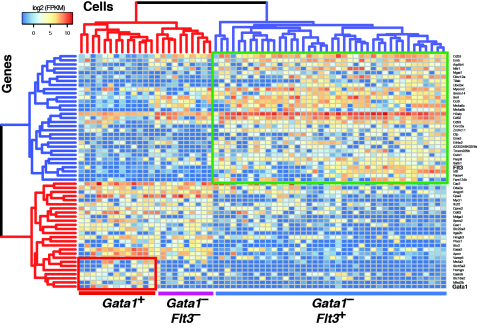
Use of single cell RNA sequencing to identify novel myeloid progenitor subsets, each column representing the gene expression of a single preGM myeloid progenitor cell. The analysis shows that the preGM population can be clustered into three subsets defined by Gata1 and Flt3 expression. These subsets were shown to define two distinct cellular pathways of myelopoiesis (Drissen et al. 2016).
In order to produce mature blood cells, HSCs generate multi-potent progenitor cells, which via a complex hierarchy of increasingly restricted progenitors, become committed to the production of a single blood cell type. We have used single cell RNA sequencing (see Figure) and single cell functional progenitor analysis to generate a new model of the hematopoietic hierarchy (Drissen et al., 2016), and are currently extending these studies to the human system. As leukemias occur when progenitors become malignant these results have paved the way for better understanding of human hematopoietic malignancies.
All of these processes are underpinned by supporting cells (stroma) that provide growth factors and cell-cell signaling to hematopoietic cells, and a key focus of the lab is to understand how changes to stromal cell populations contribute to both hematopoietic aging and leukemia progression. We recently identified vascular endothelial cells as an important stromal cell in T-cell development (Buono et al. 2016; Figure), and are currently using high-throughput single cell profiling of stromal populations to find novel cell populations and to identify the changes that occur to the bone marrow stroma during ageing and when leukemia develops.
DPHIL PROJECTS AVAILABLE: